Metode berhirarki
Ref: https://rpubs.com/odenipinedo/cluster-analysis-in-R
library(readxl)
Provinsi <- read_excel("Data/provinsi.xlsx")
Prov.scaled = scale(Provinsi[,c(4:8)])
rownames(Prov.scaled) = Provinsi$Provinsi
head(Prov.scaled)
IPM UHH RLS PPK Gini
Aceh 0.20822137 0.04044611 0.7444782 -0.62264434 -0.8089350
Sumut 0.20085709 -0.39282591 1.0245728 -0.11277090 -0.6516207
Sumbar 0.36532598 -0.23835502 0.4747574 0.01481560 -1.2546590
Riau 0.50033775 0.59428078 0.5162529 0.19012890 -0.9138112
Jambi 0.05848104 0.50762638 -0.1165536 -0.18648754 -0.6778397
Sumsel -0.21890679 -0.08765171 -0.2825356 -0.02582306 0.1349510
## membuat dissimilarity matrix
dprov = dist(Prov.scaled, method="euclidean")
c.comp = hclust(dprov, method = "complete")
cor(dprov , cophenetic(c.comp))
c.sing = hclust(dprov, method = "single")
cor(dprov , cophenetic(c.sing))
c.avrg = hclust(dprov, method = "average")
cor(dprov , cophenetic(c.avrg))
c.ward = hclust(dprov, method = "ward.D")
cor(dprov , cophenetic(c.ward))
c.ctrd = hclust(dprov, method = "centroid")
cor(dprov , cophenetic(c.ctrd))
library(factoextra)
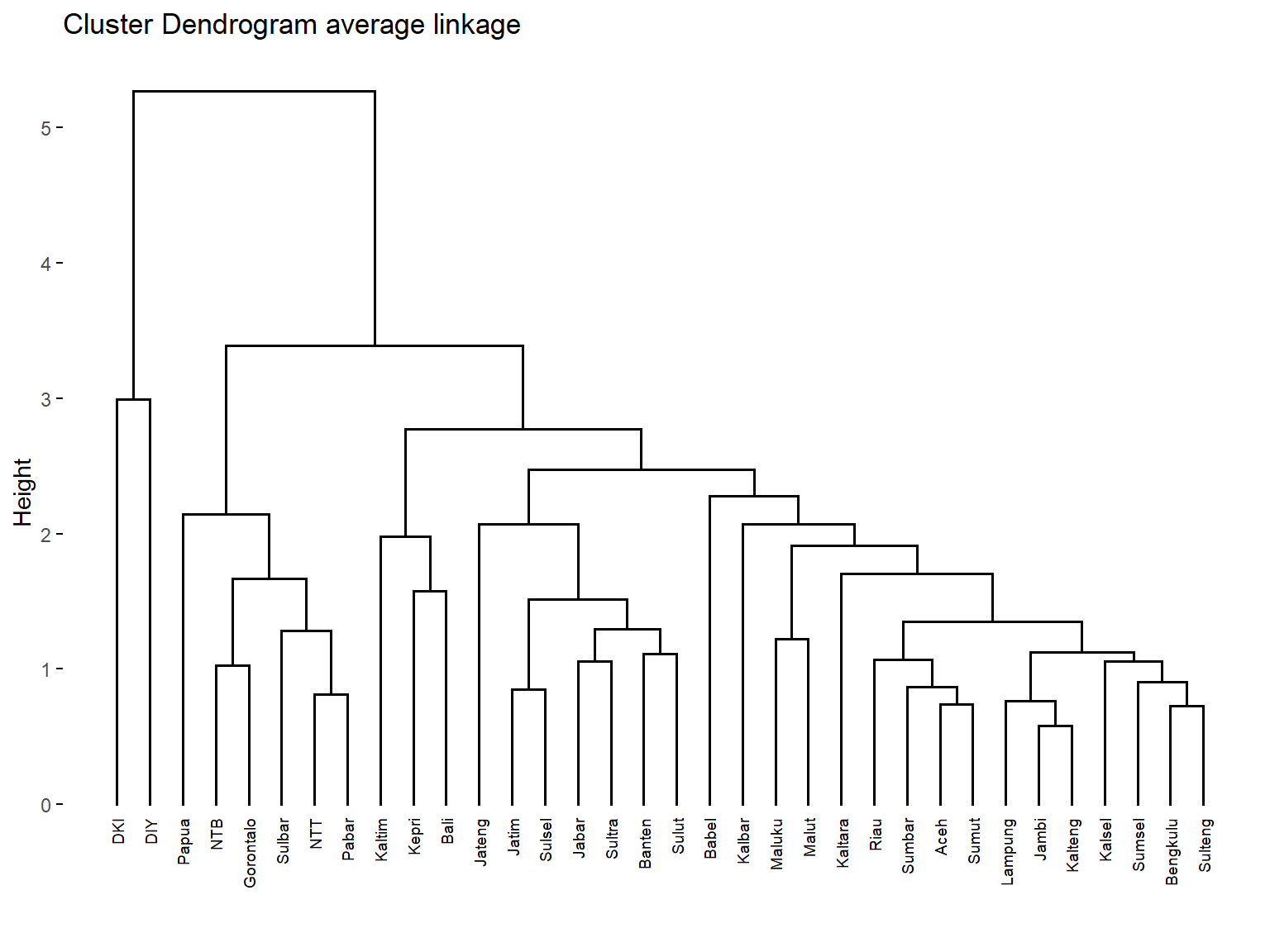
fviz_dend(c.avrg, cex = 0.5,
main = "Cluster Dendrogram average linkage")
avg_coph <- cophenetic(c.avrg)
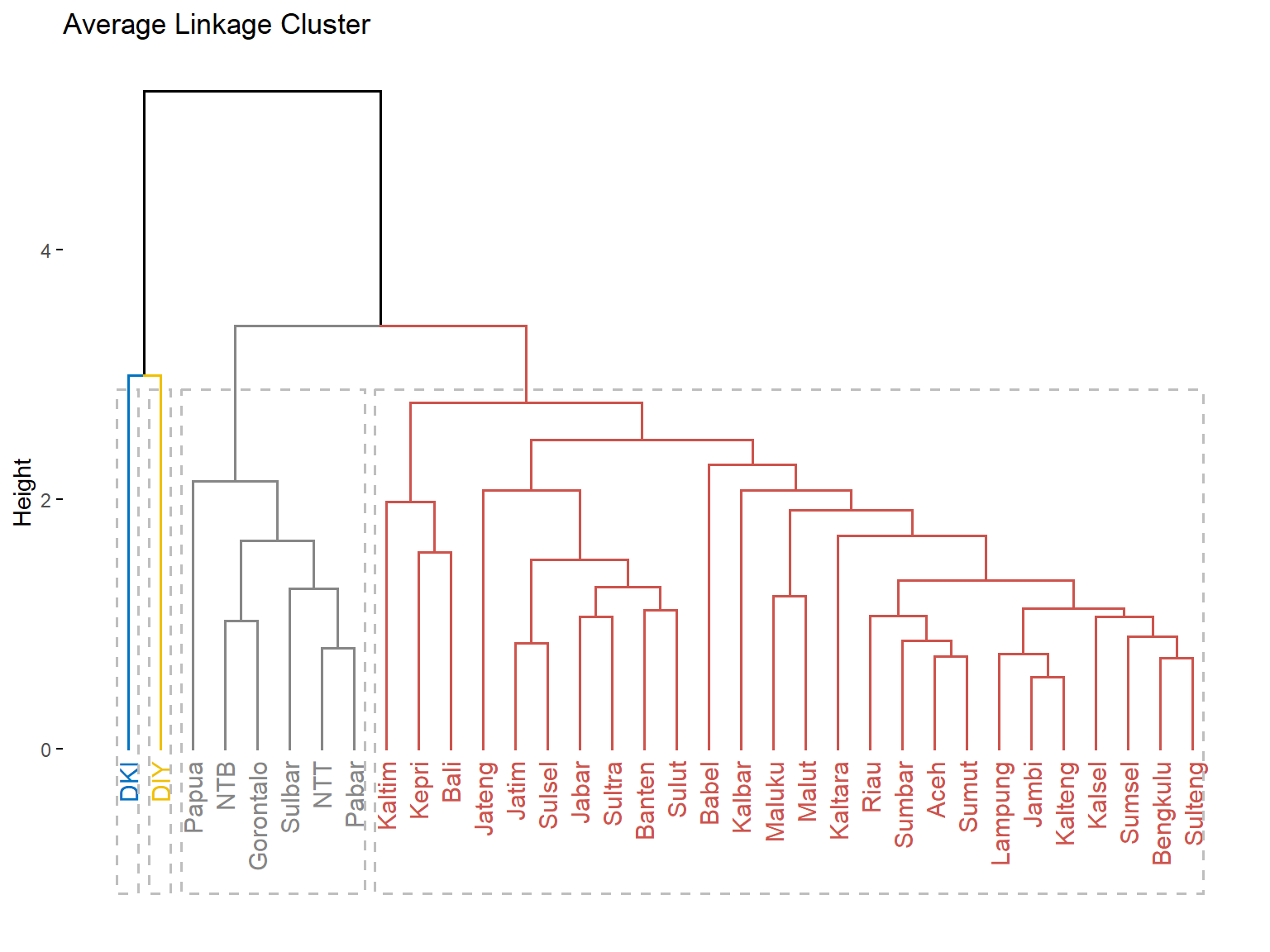
avg_clust <- cutree(c.avrg, k = 4)
table(avg_clust)
avg_clust
1 2 3 4
26 1 1 6
fviz_dend(c.avrg, k = 4,
k_colors = "jco",
rect = T,
main = "Average Linkage Cluster")
library(clValid)
library(cluster)
# internal measures
internal <- clValid(Prov.scaled, nClust = 2:6,
clMethods = "hierarchical",
validation = "internal",
metric = "euclidean",
method = "average")
summary(internal)
Clustering Methods:
hierarchical
Cluster sizes:
2 3 4 5 6
Validation Measures:
2 3 4 5 6
hierarchical Connectivity 4.5246 10.3012 11.6345 18.3198 24.1508
Dunn 0.3637 0.3703 0.3703 0.3224 0.3592
Silhouette 0.4915 0.3484 0.3092 0.2567 0.3117
Optimal Scores:
Score Method Clusters
Connectivity 4.5246 hierarchical 2
Dunn 0.3703 hierarchical 3
Silhouette 0.4915 hierarchical 2
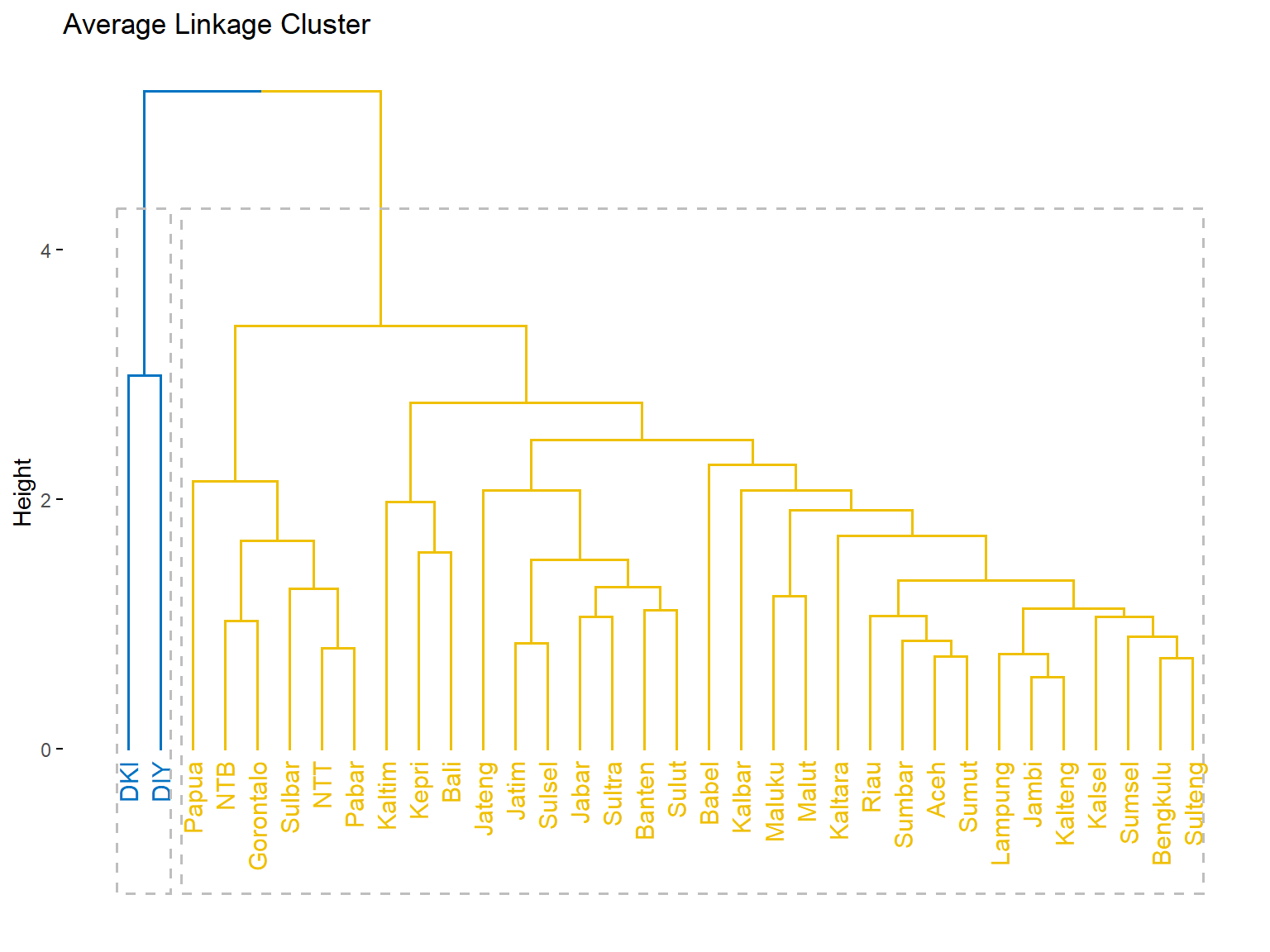
fviz_dend(c.avrg, k = 2,
k_colors = "jco",
rect = T,
main = "Average Linkage Cluster")
group = cutree(c.avrg, k = 2)
group
Aceh Sumut Sumbar Riau Jambi Sumsel Bengkulu Lampung
1 1 1 1 1 1 1 1
Babel Kepri DKI Jabar Jateng DIY Jatim Banten
1 1 2 1 1 2 1 1
Bali NTB NTT Kalbar Kalteng Kalsel Kaltim Kaltara
1 1 1 1 1 1 1 1
Sulut Sulteng Sulsel Sultra Gorontalo Sulbar Maluku Malut
1 1 1 1 1 1 1 1
Pabar Papua
1 1
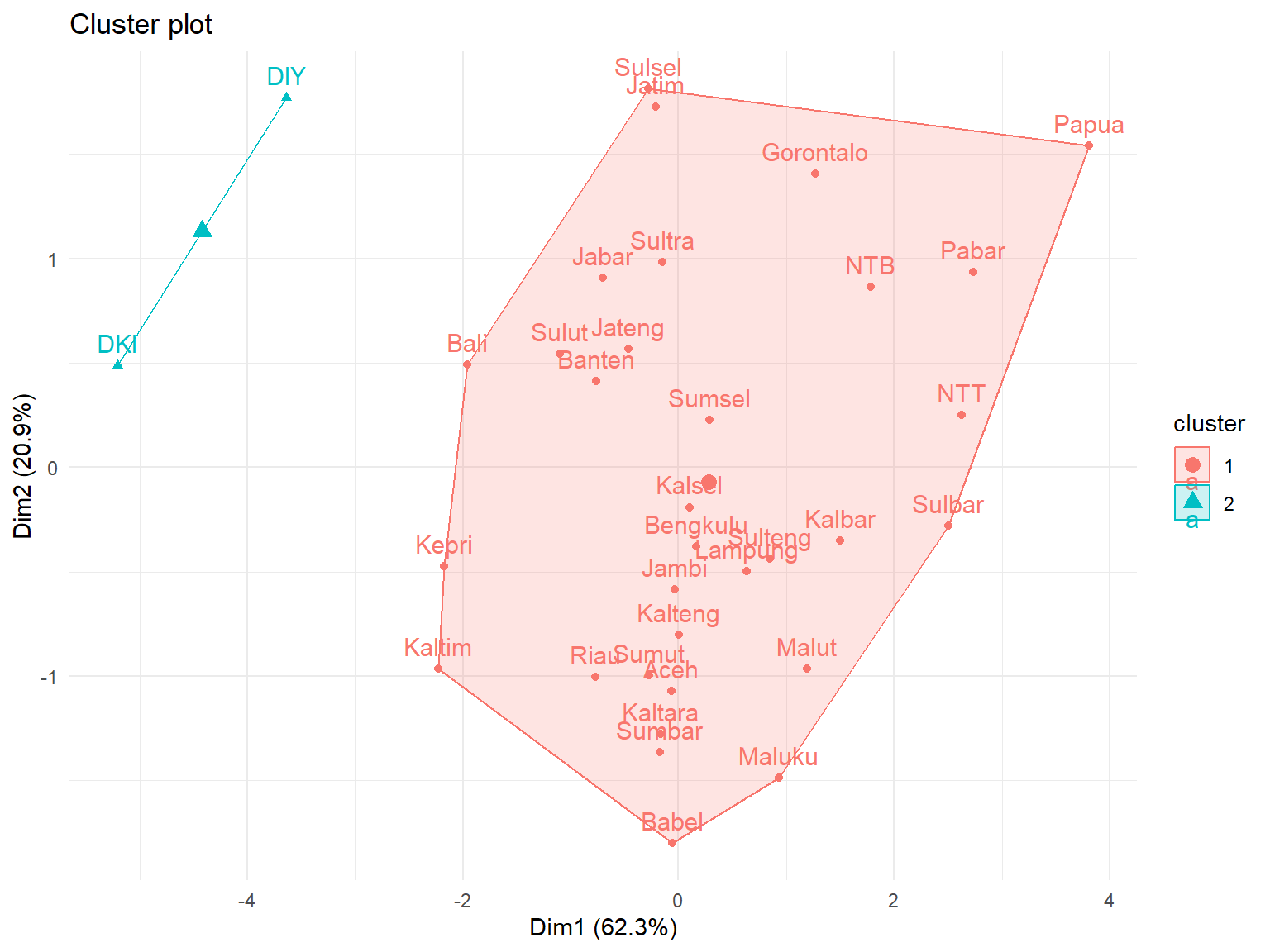
fviz_cluster(list(data = Prov.scaled,
cluster = group)) +
theme_minimal()
Standard deviations (1, .., p=5):
[1] 1.7653705 1.0227284 0.7270850 0.5299864 0.1671984
Rotation (n x k) = (5 x 5):
PC1 PC2 PC3 PC4 PC5
IPM -0.5601680 -0.05311199 -0.005227509 -0.0006949187 -0.82665781
UHH -0.4513030 0.05646383 -0.811065327 0.2024129889 0.30714735
RLS -0.4591728 -0.33781331 0.497619343 0.5648220282 0.32923179
PPK -0.5069166 0.09086739 0.227624805 -0.7546468667 0.33685862
Gini -0.1213811 0.93360390 0.206658283 0.2655422416 0.02073819
Metode tidak berhirarki - kmeans
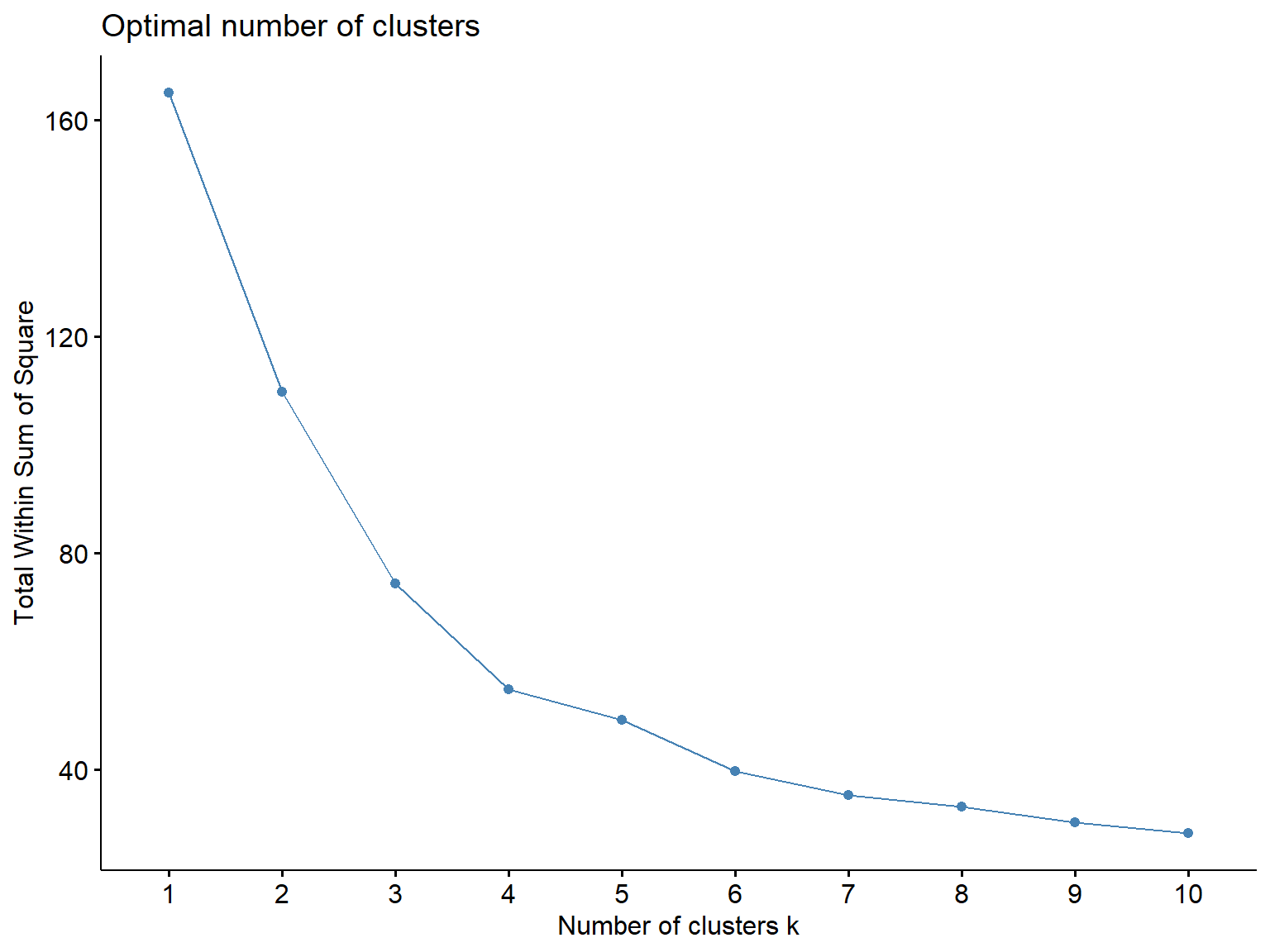
fviz_nbclust(Prov.scaled, kmeans, method = "wss")
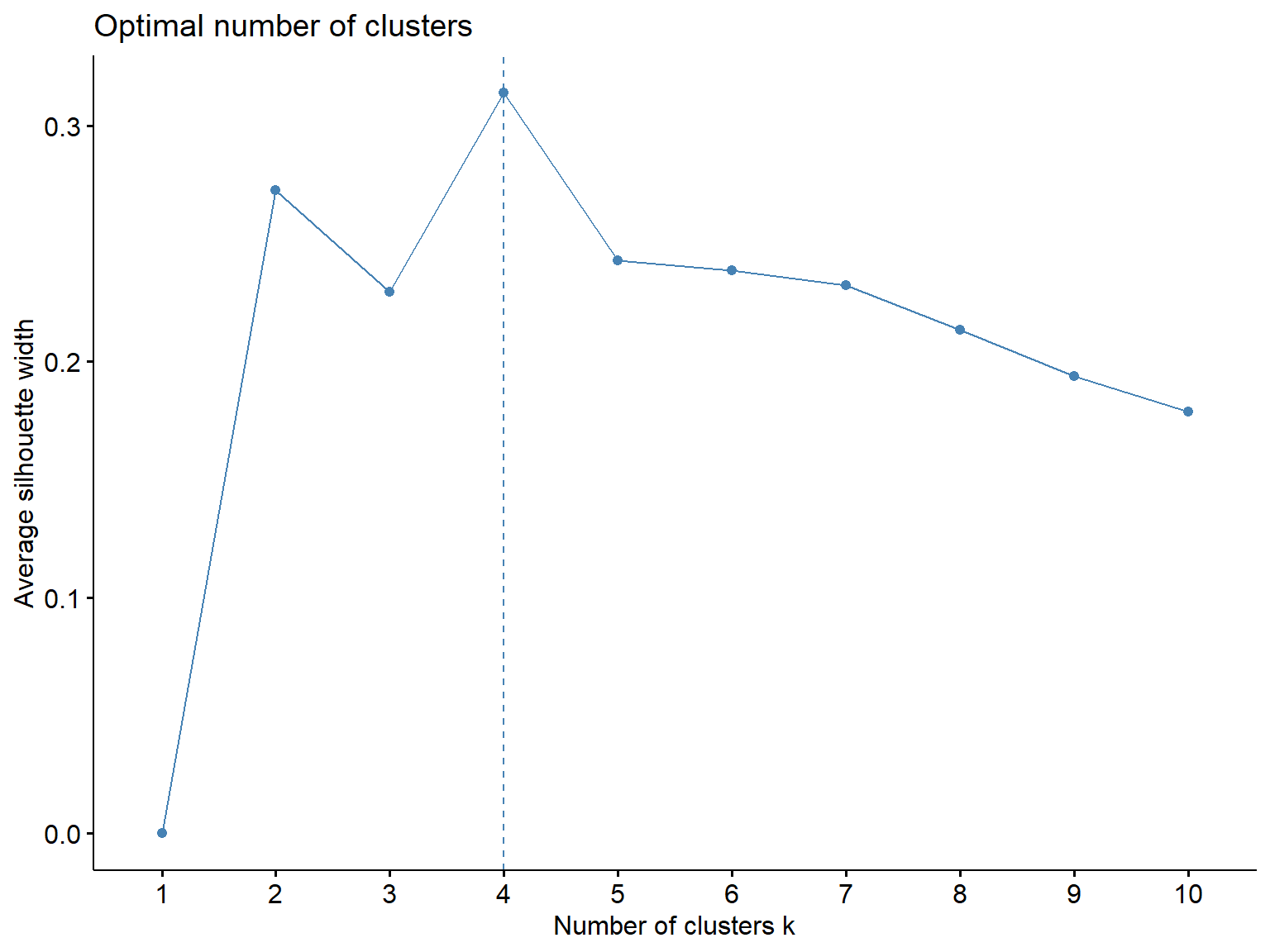
fviz_nbclust(Prov.scaled, kmeans, method = "silhouette")
set.seed(1)
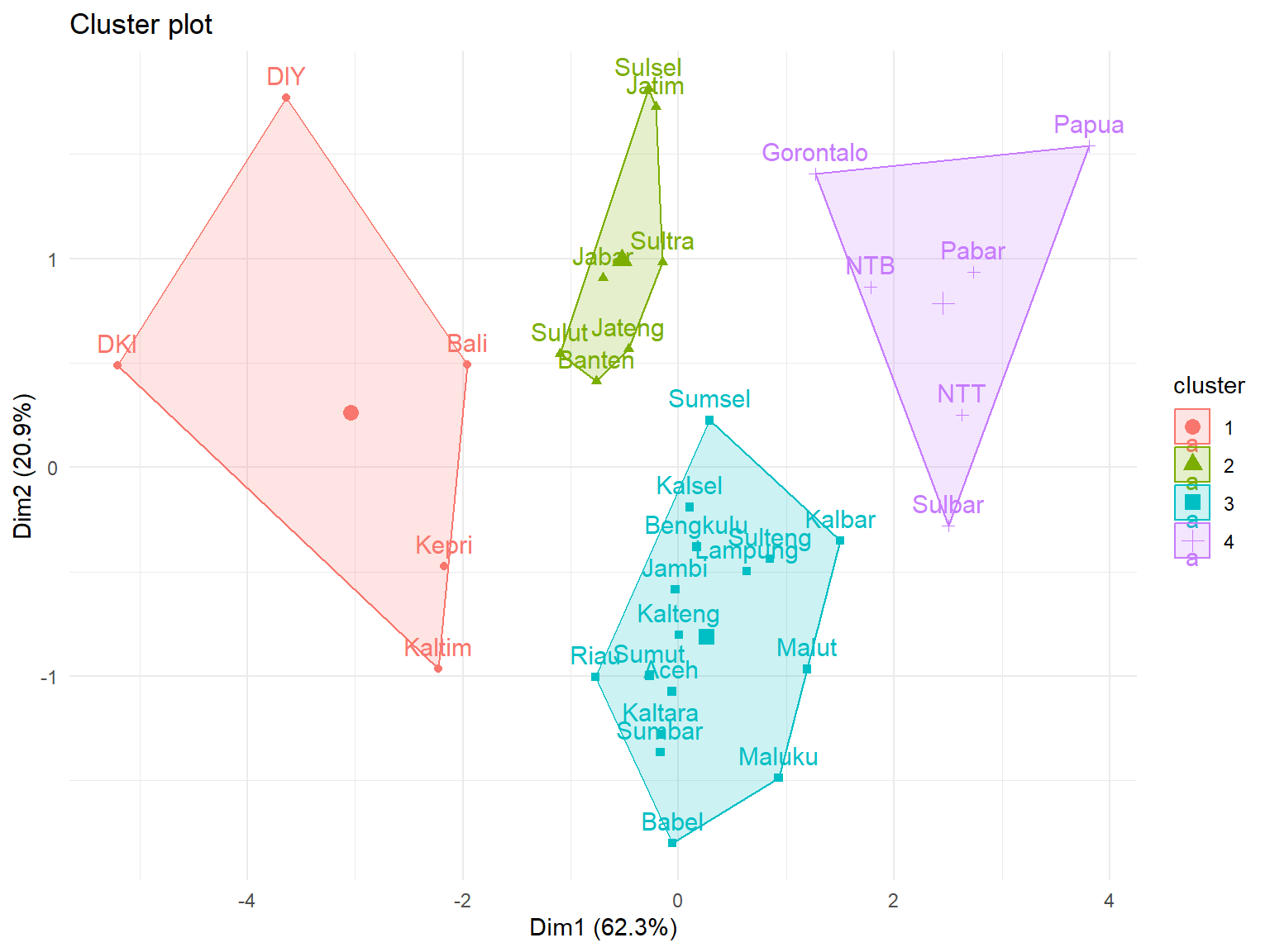
km = kmeans(Prov.scaled, centers=4)
km
K-means clustering with 4 clusters of sizes 5, 7, 16, 6
Cluster means:
IPM UHH RLS PPK Gini
1 1.67223995 1.1202353 1.3689855 1.75840321 0.6331131
2 0.22785944 0.6620973 -0.1491572 0.09292014 0.9739608
3 -0.08819085 -0.1001318 0.1246391 -0.24567350 -0.7991029
4 -1.42419372 -1.4389581 -1.2991755 -0.91861351 0.4670591
Clustering vector:
Aceh Sumut Sumbar Riau Jambi Sumsel Bengkulu Lampung
3 3 3 3 3 3 3 3
Babel Kepri DKI Jabar Jateng DIY Jatim Banten
3 1 1 2 2 1 2 2
Bali NTB NTT Kalbar Kalteng Kalsel Kaltim Kaltara
1 4 4 3 3 3 1 3
Sulut Sulteng Sulsel Sultra Gorontalo Sulbar Maluku Malut
2 3 2 2 4 4 3 3
Pabar Papua
4 4
Within cluster sum of squares by cluster:
[1] 17.054859 7.933134 22.111511 7.711994
(between_SS / total_SS = 66.8 %)
Available components:
[1] "cluster" "centers" "totss" "withinss" "tot.withinss"
[6] "betweenss" "size" "iter" "ifault"
fviz_cluster(list(data = Prov.scaled, cluster = km$cluster)) + theme_minimal()